DBT (or Data Build Tool) is a modern data transformation tool born in the cloud/DevOps era. It's a great project which much has been written about; I will try to give as brief an overview as possible.
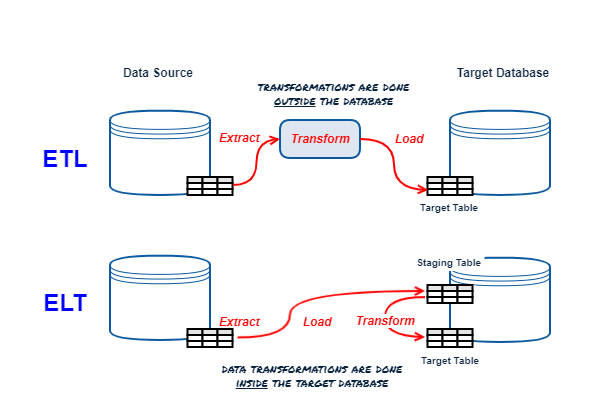
ETL vs ELT Refresher
Summary
DBT is an open-source command line tool written in Python from DBT Labs (formerly Fishtown Analytics).
DBT is designed to manage data transformations while applying software engineering best practices (including version control, automated testing, reviews, approvals, etc). Its modern software engineering and cloud-first design goals separate it from its old-school ETL/ELT predecessors.
DBT is an ELT tool focusing on the T(ransform) only, the E(xtract) and L(oad) are up to you (there are plenty of tools that specialize in this).
At its core DBT is a templating engine using Jinja (Python templating engine); it generates templates that represent SQL commands to create, replace or update objects in your database (the “T” in ELT), then oversees the execution of the templated commands. The work is "pushed down" to the underlying database containing the source and target objects and data.
Models
The concept most integral to DBT is the Model. A Model is simply a representation of a transform (or set of transforms) to a dataset, resulting in a target object (which could be a table or tables in a datamart). A model is expressed as a SELECT statement stored in a .sql file in your dbt project (well get to that in a minute).
Suppose we want to create a denormalized fact table for commits to store in a datamart in BigQuery. This is what a model file might look like (using the BigQuery SQL dialect and referencing objects that should exist and be accessible at runtime when we execute the model).
{{ config(materialized='table') }}
with commits as (
SELECT
SUBSTR(commit, 0, 13) as commit_short_sha,
committer.name as commiter_name,
committer.date as commit_date,
message,
repo_name
FROM `bigquery-public-data.github_repos.sample_commits` c
)
select * from commits
Models are created as views by default, but you can materialize these as tables where needed.
You configure connectivity to the target database using adapters (software libraries provided by dbt in the case of most mainstream databases) and profiles (which contain details around authentication, databases/datasets, schemas etc).
DBT Project
A DBT project is simply a folder containing your models and some other configuration data. You can initialize this by running dbt init from your desired project directory. In its most basic form, the structure looks like this:
models/
├─ your_model.sql
├─ schema.yml
dbt_project.yml
Your models can be created under subfolders for organization. schema.yml is an optional file that contains tests for columns, can also include descriptions for documentation. The dbt_project.yml file is the main entry point for the dbt program, it contains the configuration for the project, including which profile to use. Profiles (stored in a file called profiles.yml store all of the necessary connectivity information for your target database platform. By default dbt init creates this file a .dbt folder under your home directory.
You could store this with your project (be careful not to commit secrets like database credentials to source control). If you store it in any other directory than the default, you will need to tell dbt where it can find this file using the --profiles-dir argument of any dbt command, see here for more information.
To confirm your project is ship shape, run dbt parse; if there are no errors, you are good to proceed running and testing your models.
Running DBT Models
To run your models, simply run the following command from the directory containing your dbt_project.yml file (typically the root of your project folder):
- Command
- Output
06:56:36 Running with dbt=1.2.0
06:56:36 Found 1 model, 2 tests, 0 snapshots, 0 analyses, 285 macros, 0 operations, 0 seed files, 0 sources, 0 exposures, 0 metrics
06:56:36
06:56:37 Concurrency: 1 threads (target='dev')
06:56:37
06:56:37 1 of 1 START table model dbt_dataset.fct_commits ............................... [RUN]
06:56:50 1 of 1 OK created table model dbt_dataset.fct_commits .......................... [CREATE TABLE (672.3k rows, 396.7 MB processed) in 13.28s]
06:56:50
06:56:50 Finished running 1 table model in 0 hours 0 minutes and 14.01 seconds (14.01s).
06:56:50
06:56:50 Completed successfully
06:56:50
06:56:50 Done. PASS=1 WARN=0 ERROR=0 SKIP=0 TOTAL=1Model deployed! Let's test it:
- Command
- Output
06:57:19 Running with dbt=1.2.0
06:57:19 Found 1 model, 2 tests, 0 snapshots, 0 analyses, 285 macros, 0 operations, 0 seed files, 0 sources, 0 exposures, 0 metrics
06:57:19
06:57:19 Concurrency: 1 threads (target='dev')
06:57:19
06:57:19 1 of 2 START test not_null_fct_commits_commit_short_sha ........................ [RUN]
06:57:21 1 of 2 PASS not_null_fct_commits_commit_short_sha .............................. [PASS in 1.88s]
06:57:21 2 of 2 START test unique_fct_commits_commit_short_sha .......................... [RUN]
06:57:24 2 of 2 PASS unique_fct_commits_commit_short_sha ................................ [PASS in 3.14s]
06:57:24
06:57:24 Finished running 2 tests in 0 hours 0 minutes and 5.41 seconds (5.41s).
06:57:24
06:57:24 Completed successfully
06:57:24
06:57:24 Done. PASS=2 WARN=0 ERROR=0 SKIP=0 TOTAL=2This will run all of the tests associated with your model(s) - in this case, not null and unique tests defined in the schema.yml file. That's it, deployed and tested.
Other Stuff
There is some other stuff in DBT you should be aware of, like seeds, snapshots, analyses, macros, and more but our five minutes is up 😃. We can discuss these next time; you are up and running with the basics of DBT now, get transforming!
if you have enjoyed this post, please consider buying me a coffee ☕ to help me keep writing!