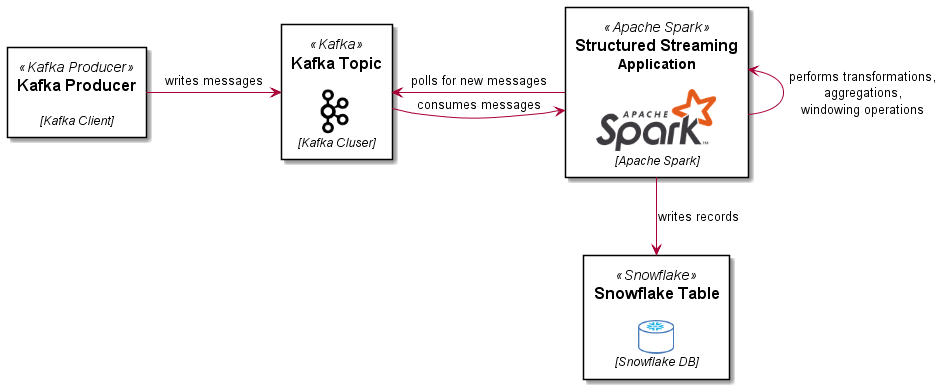
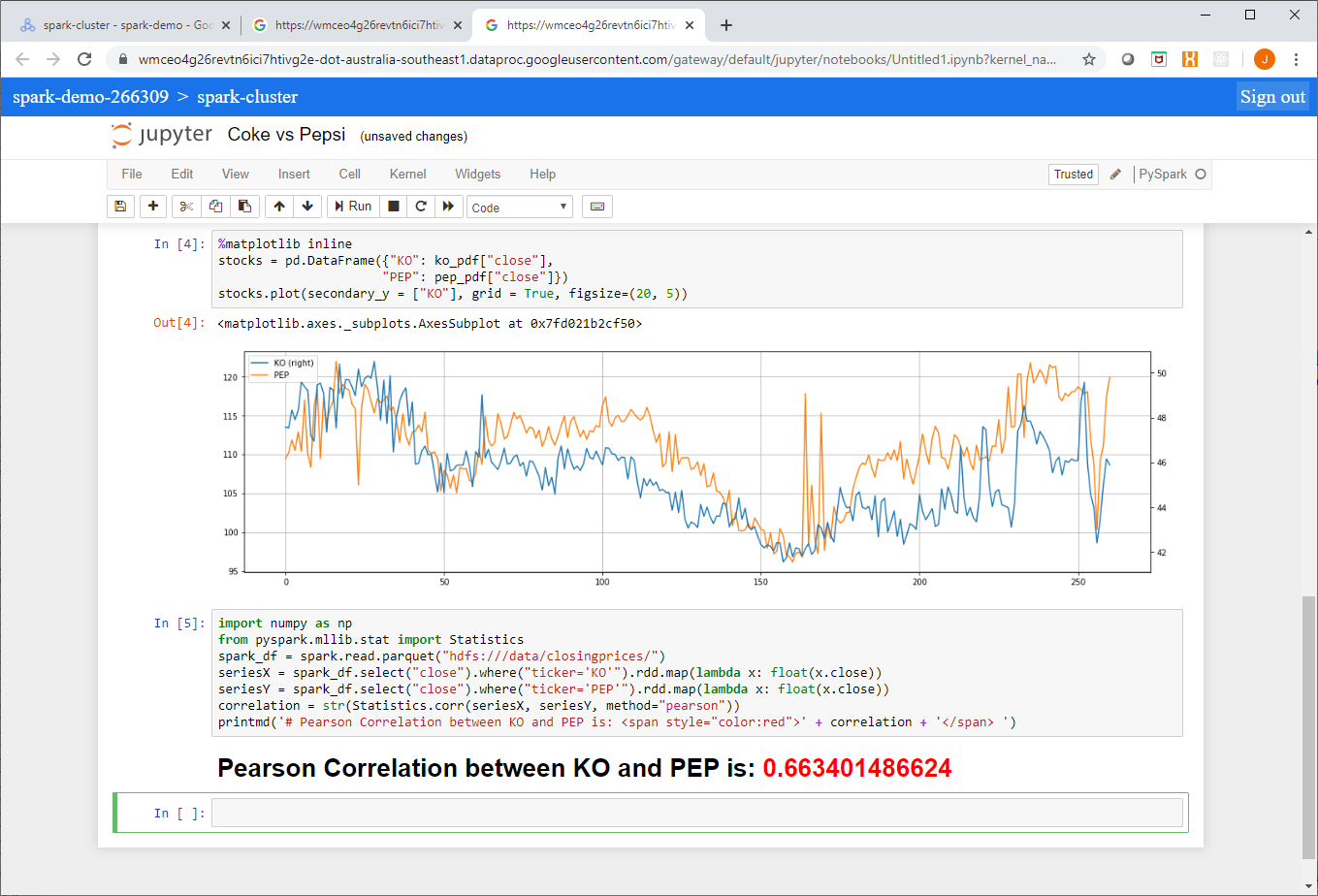
Loading Parquet format files into BigQuery is straightforward, you just need to specify the file location (local, Google Cloud Storage, Drive, Amazon S3 or Azure Blob storage) and thats pretty much it, BigQuery works the rest out from there.
bq load \
--location=australia-southeast2 \
--project_id=parquet-demo \
--source_format=PARQUET \
parquet_test.dim_calendar \
.\Calendar.gzip
In Snowflake, however, it is not as simple, I'll share my approach to automating this here.
Parquet is a self-describing, column-oriented storage format commonly used in distributed systems for input and output. Data in Parquet files is serialised for optimised consumption from Parquet client libraries and packages such as pandas, pyarrow, fastparquet, dask, and pyspark.
Background
Data in a Parquet file is stored in a single column for a self-contained dataset. If you were to ingest this into Snowflake without knowing the schema you could do something like this...
CREATE OR REPLACE TABLE PARQUET_TEST.PUBLIC.DIM_CALENDAR (
Data variant
);
COPY INTO PARQUET_TEST.PUBLIC.DIM_CALENDAR
(
Data
) FROM (
SELECT
*
FROM
@PARQUET_TEST.PUBLIC.DIM_CALENDAR_STAGE)
file_format = (TYPE = parquet);
You would end up with something like...
Row | Data |
|---|---|
1 | {"CalMonthOfYearNo": 6, "CalYear": 2020, ... } |
2 | {"CalMonthOfYearNo": 6, "CalYear": 2020, ... } |
... | ... |
You could then have a second stage of processing to convert this into a normal relational structure.
Or you could do this in one step, with a little prep work ahead of time. In my scenario I was given several parquet files from a client for a one-off load into Snowflake, several files for a fact table and multiple single files representing different dimension tables.
Streamlined Ingestion for Parquet Files into Snowflake
To collapse the formatting and uploading of Parquet files into a materialized table into one step, we need to do a couple of things:
- Create the target table with the correct schema (column names and data types); and
- perform a projection in our
COPYcommand from the single column containing all of the data (represented by$1in Snowflake) into columns defined in step 1
Since this is technically a transformation and only named stages are supported for COPY transformations, we need to create a stage for the copy. In my case there is a pre-existing Storage Integration in place that can be used by the stage.
Generate Table DDL
To automate the generation of the DDL to create the table and stage and the COPY command, I used Python and Spark (which has first class support for Parquet files). Parquet datatypes are largely the same as Snowflake, but if we needed to, we could create a map and modify the target types during the DDL generation.
First copy specimen Parquet formatted files to a local directory, the script we are creating can then iterate through the parquet files and generate all of the commands we will need saved to a .sql file.
With some setup information provided (not shown for brevity), we will first go through each file in the directory, capture metadata along with the schema (column name and data type) as shown here:
for file in files:
tableMap = {}
table = file.stem
spark = launch_spark_session()
parquetFile = spark.read.parquet("%s/%s" %(BASE_DIR, file))
data_types = parquetFile.dtypes
stop_spark_session(spark)
tableMap['name'] = table
tableMap['file'] = file
tableMap['data_types'] = data_types
allTables.append(tableMap)
The allTables list looks something like this...
[{'name': 'Calendar', 'file': PosixPath('data/dim/Calendar.gzip'), 'data_types': [('Time_ID', 'bigint'), ('CalYear', 'bigint'), ('CalMonthOfYearNo', 'bigint'), ('FinYear', 'bigint'), ('FinWeekOfYearNo', 'bigint')]}, ... ]
Next we generate the CREATE TABLE statement using the allTables list:
# create output file for all sql
with open('all_tables.sql', 'w') as f:
for table in allTables:
print("processing %s..." % table['name'])
f.write("/*** Create %s Table***/" % table['name'].upper())
sql = """
CREATE OR REPLACE TABLE %s.%s.%s (
""" % (database, schema, table['name'].upper())
for column in table['data_types']:
sql += " %s %s,\n" % (column[0], column[1])
sql = sql[:-2] + "\n);"
f.write(sql)
f.write("\n\n")
Generate Named Stage DDL
Then we generate the stage in S3 from which the files will be loaded:
f.write("/*** Create %s Stage***/" % table['name'].upper())
sql = """
CREATE OR REPLACE STAGE %s.%s.%s_STAGE
url='%s/%s'
storage_integration = %s
encryption=(type='AWS_SSE_KMS' kms_key_id = '%s');
""" % (database, schema, table['name'].upper(), s3_prefix, table['file'], storage_int, kms_key_id)
f.write(sql)
f.write("\n\n")
Generate COPY commands
Then we generate the COPY commands...
f.write("/*** Copying Data into %s ***/" % table['name'].upper())
sql = """
COPY INTO %s.%s.%s
(\n""" % (database, schema, table['name'].upper())
for column in table['data_types']:
sql += " %s,\n" % column[0]
sql = sql[:-2] + "\n)"
sql += " FROM (\nSELECT\n"
for column in table['data_types']:
sql += " $1:%s::%s,\n" % (column[0], column[1])
sql = sql[:-2] + "\nFROM\n"
sql += "@%s.%s.%s_STAGE)\n" % (database, schema, table['name'].upper())
sql += " file_format = (TYPE = parquet);"
f.write(sql)
f.write("\n\n")
Since this is a one off load, we will go ahead and drop the stage we created as it is no longer needed (this step is optional)..
f.write("/*** Dropping stage for %s ***/" % table['name'].upper())
sql = """
DROP STAGE %s.%s.%s_STAGE;
""" % (database, schema, table['name'].upper())
f.write(sql)
f.write("\n\n")
The resultant file created looks like this..
/*** Create CALENDAR Table***/
CREATE OR REPLACE TABLE PARQUET_TEST.PUBLIC.DIM_CALENDAR (
Time_ID bigint,
CalYear bigint,
CalMonthOfYearNo bigint,
FinYear bigint,
FinWeekOfYearNo bigint
);
/*** Create DIM_CALENDAR Stage***/
CREATE OR REPLACE STAGE PARQUET_TEST.PUBLIC.DIM_CALENDAR_STAGE
url='s3://my-bucket/data/dim/Calendar.gzip'
storage_integration = my_storage_int
encryption=(type='AWS_SSE_KMS' kms_key_id = '4f715ec9-ee8e-44ab-b35d-8daf36c05f19');
/*** Copying Data into DIM_CALENDAR ***/
COPY INTO PARQUET_TEST.PUBLIC.DIM_CALENDAR
(
Time_ID,
CalYear,
CalMonthOfYearNo,
FinYear,
FinWeekOfYearNo
) FROM (
SELECT
$1:Time_ID::bigint,
$1:CalYear::bigint,
$1:CalMonthOfYearNo::bigint,
$1:FinYear::bigint,
$1:FinWeekOfYearNo::bigint
FROM
@PARQUET_TEST.PUBLIC.DIM_CALENDAR_STAGE)
file_format = (TYPE = parquet);
/*** Dropping stage for DIM_CALENDAR ***/
DROP STAGE PARQUET_TEST.PUBLIC.DIM_CALENDAR_STAGE;
Load your data
You can then run this along with all of the other dimension and fact table DDL and COPY commands generated to perform the one-off load from parquet files. You can find the complete code below, enjoy!
Complete Code
from pathlib import Path
from pyspark.sql import SparkSessiondef launch_spark_session():
return SparkSession \
.builder \
.appName("Parquet DDL Generation") \
.getOrCreate()
def stop_spark_session(spark):
spark.stop()
allTables = []
database = "PARQUET_TEST"
schema = "PUBLIC"
s3_prefix = 's3://my-bucket'
storage_int = 'my_storage_int'
kms_key_id = '4f715ec9-ee8e-44ab-b35d-8daf36c05f19'
BASE_DIR = Path(__file__).resolve().parent
directory = 'data/dim'
files = Path(directory).glob('*.gzip')
for file in files:
tableMap = {}
table = file.stem
spark = launch_spark_session()
parquetFile = spark.read.parquet("%s/%s" %(BASE_DIR, file))
data_types = parquetFile.dtypes
stop_spark_session(spark)
tableMap['name'] = table
tableMap['file'] = file
tableMap['data_types'] = data_types
allTables.append(tableMap)
# create output file for all sql
with open('all_tables.sql', 'w') as f:
for table in allTables:
print("processing %s..." % table['name'])
f.write("/*** Create %s Table***/" % table['name'].upper())
sql = """
CREATE OR REPLACE TABLE %s.%s.%s (
""" % (database, schema, table['name'].upper())
for column in table['data_types']:
sql += " %s %s,\n" % (column[0], column[1])
sql = sql[:-2] + "\n);"
f.write(sql)
f.write("\n\n")
f.write("/*** Create %s Stage***/" % table['name'].upper())
sql = """
CREATE OR REPLACE STAGE %s.%s.%s_STAGE
url='%s/%s'
storage_integration = %s
encryption=(type='AWS_SSE_KMS' kms_key_id = '%s');
""" % (database, schema, table['name'].upper(), s3_prefix, table['file'], storage_int, kms_key_id)
f.write(sql)
f.write("\n\n")
f.write("/*** Copying Data into %s ***/" % table['name'].upper())
sql = """
COPY INTO %s.%s.%s
(\n""" % (database, schema, table['name'].upper())
for column in table['data_types']:
sql += " %s,\n" % column[0]
sql = sql[:-2] + "\n)"
sql += " FROM (\nSELECT\n"
for column in table['data_types']:
sql += " $1:%s::%s,\n" % (column[0], column[1])
sql = sql[:-2] + "\nFROM\n"
sql += "@%s.%s.%s_STAGE)\n" % (database, schema, table['name'].upper())
sql += " file_format = (TYPE = parquet);"
f.write(sql)
f.write("\n\n")
f.write("/*** Dropping stage for %s ***/" % table['name'].upper())
sql = """
DROP STAGE %s.%s.%s_STAGE;
""" % (database, schema, table['name'].upper())
f.write(sql)
f.write("\n\n")
if you have enjoyed this post, please consider buying me a coffee ☕ to help me keep writing!