Container images provide an ideal software packaging solution for DataOps and python based data pipeline workloads. Containers enable Data Scientists and Data Engineers to incorporate the latest packages and libraries without the issues associated with introducing breaking changes into shared environments. A Data Engineer or Data Scienctist can quickly release new functionality with the best tools available.
Container images provide safer developer environments but as the number of container images used for production workloads grow, a maintenance challenge can emerge. Whether using pip or poetry to manage python packages and dependencies, updating a container definition requires edits to the explicit package versions as well as to the pinned or locked versions of the package dependencies. This process can be error prone without automation and a repeatable CICD workflow.
A workflow pattern based on docker buildkit / moby buildkit multi-stage builds provides an approach that maintains all the build specifications in a single Dockerfile, while build tools like make provide a simple and consistent interface into the container build stages. The data pipeline challenges addresses with a multi-stage build pattern include:
- automating lifecycle management of the Python packages used by data pipelines
- integrating smoke testing of container images to weed out compatibility issues early
- simplifying the developer experience with tools like
make that can be used both locally and in CI/CD pipelines
The Dockerfile contains the definitions of the different target build stages and order of execution from one stage to the next. The Makefile wraps the Dockerfile build targets into a standard set of workflow activities, following a similar to $ config && make && make install
A typical dataops/gitops style workflow for maintaining container images includes actions in the local environment to define the required packages and produce the pinned dependency poetry.lock file or requirements.txt packages list containing the full set of pinned dependent packages.
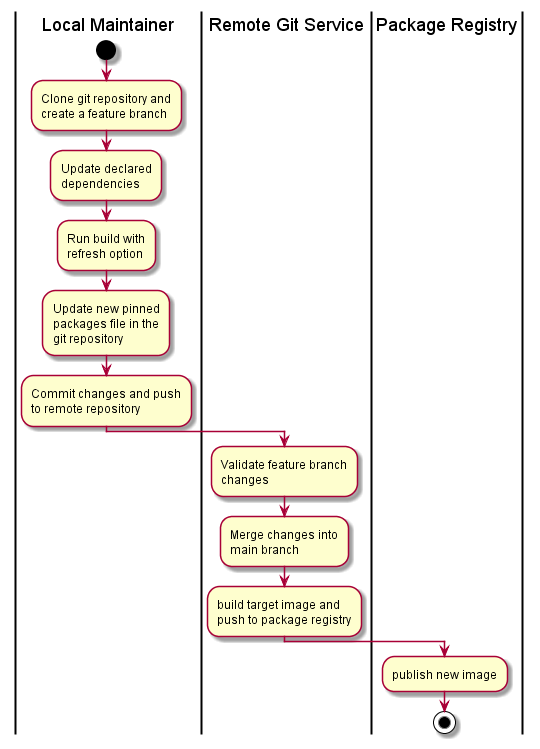
Given and existing project in a remote git repository with a CI/CD pipeline defined, the following workflow would be used to update package versions and dependencies:
@startuml Multi-stage build workflow
|Local Maintainer|
start
:Clone git repository and
create a feature branch;
:Update declared
dependencies;
:Run build with
refresh option;
:Update new pinned
packages file in the
git repository;
:Commit changes and push
to remote repository;
|Remote Git Service|
:Validate feature branch
changes;
:Merge changes into
main branch;
:build target image and
push to package registry;
|Package Registry|
:publish new image;
stop
@enduml
The image maintainer selects the packages to update or refresh using a local development environment, working from a feature branch. This includes performing an image smoke-test to validate the changes within the container image.
Once refreshed image has been validated, the lock file or full pinned package list is commited back to the repository and pushed to the remote repository. The CI/CD pipeline performs a trial build and conducts smoke testing. On merge into the main branch, the target image is built, re-validated, and pushed to the container image registry.
The multi-stage build pattern can support both defining both the declared packages for an environment as well as the dependent packages, but poetry splits the two into distinct files, a pyproject.toml file containing the declated packages and a poetry.lock file that contains the full set of declared and dependent packages, including pinned versions. pip supports loading packages from different files, but requires a convention for which requirements file contains the declared packages and while contains the full set of pinned package versions produced by pip freeze. The example code repo contains examples using both pip and poetry.
The following example uses poetry in a python:3.8 base image to illustrate managing the dependencies and version pinning of python packages.
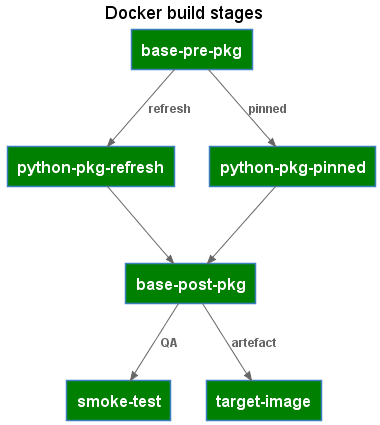
The Dockerfile defines the build stages used for both local refresh and by the CICD pipelines to build the target image.
@startuml Dockerfile stages
!define C4_PLANTUML https://raw.githubusercontent.com/plantuml-stdlib/C4-PlantUML/master
!include C4_PLANTUML/C4_Component.puml
HIDE_STEREOTYPE()
UpdateElementStyle(Container, $bgColor=green)
Title: Docker build stages
Container(pre, base-pre-pkg,)
Container(refresh, python-pkg-refresh,)
Container(pinned, python-pkg-pinned,)
Container(post, base-post-pkg,)
Container(smoke, smoke-test,)
Container(target, target-image,)
Rel(pre, refresh, "refresh")
Rel(pre, pinned, "pinned")
Rel(refresh, post, " ")
Rel(pinned, post, " ")
Rel(post, smoke, "QA")
Rel(post, target, "artefact")
@enduml
The Dockerfile makes use of the docker build arguments feature to pass in whether the build should refresh package versions or build the image from pinned packages.
Any image setup and pre-python package installation steps. For poetry, this includes setting the config option to skip the creation of a virtual environment as the container already provides the required isolation.
ARG PYTHON_PKG_VERSIONS=pinned
FROM python:3.8 as base-pre-pkg
RUN install -d /src && \
pip install --no-cache-dir poetry==1.1.13 && \
poetry config virtualenvs.create false
WORKDIR /src
The steps to generate a poetry.lock file containing the pinned package versions.
FROM base-pre-pkg as python-pkg-refresh
COPY pyproject.toml poetry.lock /src/
RUN poetry update && \
poetry install
The steps to install packages using the pinned package versions.
FROM base-pre-pkg as python-pkg-pinned
COPY pyproject.toml poetry.lock /src/
RUN poetry install
Build Stage: base-post-pkg
A consolidation build target that can refer to either the python-pkg-refresh or the python-pkg-pinned stages, depending on the docker build argument and includes any post-package installation steps.
FROM python-pkg-${PYTHON_PKG_VERSIONS} as base-post-pkg
Simple smoke tests and validation commands to validate the built image.
FROM base-post-pkg as smoke-test
WORKDIR /src
COPY tests/ ./tests
RUN poetry --version && \
python ./tests/module_smoke_test.py
The final build target container image. Listing the target-image as the last stage in the Dockerfile has the effect of also making this the default build target.
FROM base-post-pkg as target-image
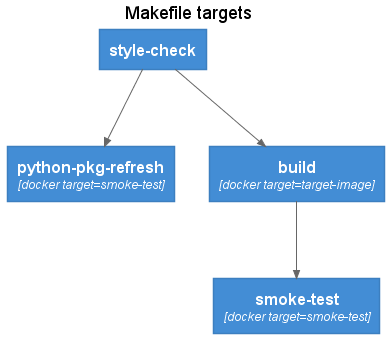
The Makefile provides a workflow oriented wrapper over the Dockerfile build stage targets. The Makefile targets can be executed both in a local development environment as well as via a CICD pipeline. The Makefile includes several variables that can either be run using default values, or overridden by the CI/CD pipeline.
@startuml Makefile targets
!define C4_PLANTUML https://raw.githubusercontent.com/plantuml-stdlib/C4-PlantUML/master
!include C4_PLANTUML/C4_Component.puml
HIDE_STEREOTYPE()
Title: Makefile targets
Container(style, style-check,)
Container(refresh, python-pkg-refresh, "docker target=smoke-test")
Container(smoke, smoke-test, "docker target=smoke-test")
Container(build, build, "docker target=target-image")
Rel(style, refresh, " ")
Rel(style, build, " ")
Rel(build, smoke, " ")
@enduml
Linting and style checking of source code. Can include both application code as well as the Dockerfile itself using tools such as hadolint.
style-check:
hadolint ./Dockerfile
The python-pkg-refresh target builds a version of the target image with refreshed package versions. A temporary container instance is created from the target image and the poetry.lock file is copied into the local file system. The smoke-test docker build target is used to ensure image validation is also performed.
The temporary container as well as the package refresh image are removed after the build.
python-pkg-refresh:
@echo ">> Update python packages in container image"
docker build ${DOCKER_BUILD_ARGS} \
--target smoke-test \
--build-arg PYTHON_PKG_VERSIONS=refresh \
--tag ${TARGET_IMAGE_NAME}:$@ .
@echo ">> Copy the new poetry.lock file with updated package versions"
docker create --name ${TARGET_IMAGE_NAME}-$@ ${TARGET_IMAGE_NAME}:$@
docker cp ${TARGET_IMAGE_NAME}-$@:/src/poetry.lock .
@echo ">> Clean working container and refresh image"
docker rm ${TARGET_IMAGE_NAME}-$@
docker rmi ${TARGET_IMAGE_NAME}:$@
The standard build target using pinned python package versions.
build:
docker build ${DOCKER_BUILD_ARGS} \
--target target-image \
--tag ${TARGET_IMAGE_NAME}:${BUILD_TAG} .
Builds an image and peforms smoke testing. The smoke-testing image is removed after the build.
smoke-test:
docker build ${DOCKER_BUILD_ARGS} \
--target smoke-test \
--tag ${TARGET_IMAGE_NAME}:$@ .
docker rmi ${TARGET_IMAGE_NAME}:$@
The toolchain combination of multi-stage container image builds with make provides a codified method for the lifecycle management of the containers used in data science and data engineering workloads.
The maintainer:
git checkout -b my-refresh-feature
make python-pkg-refresh
make smoke-test
git add pyproject.toml poetry.lock
git commit -m "python package versions updated"
git push
The CICD pipeline:
make build
make smoke-test
docker push <target-image>:<build-tag>